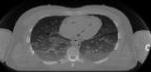
Exercise 1: Ultrasound image display
Monday, September 9, 14.30-17.00 in the
E-databar, build 341, ground floor, room 015.
Purpose:
The purpose of this exercise is to demonstrate the signal processing involved
in ultrasound image display. This includes finding the enveloped through
a Hilbert transform, compressing the data, and making the image interpolation.
It also show how serveral frames can be combined into one movie.
Preparation:
Read Chapter 2 in the book: Jørgen Arendt Jensen: Estimation
of Blood Velocities Using Ultrasound, A Signal Processing Approach, Cambridge
University Press, 1996.
Go through the different exercise points and write down suggestions for
your Matlab code.
Hints using Matlab:
Write all Matlab instructions in a file (f. ex. ex1.m) and run the file
in the Matlab window by typing ex1 at the Matlab prompt. With this approach
you will be able to receive help from the instructor about problems with
your code.
The data files can also been found on
DTU Inside under exercise1. This can be used, if you have problems downloading the
files from this page.
Exercise:
-
Log in to one of the PCs. Make a directory course_31545,
where all the course files can be stored.
-
Start a web browser and go to the course home page at:
https://courses.healthtech.dtu.dk/22485
Go to exercise1 (this page at:
https://courses.healthtech.dtu.dk/22485/?exercises/exercise1/exercise1.html)
and download the image data by right-clicking
here. Place the datafile with the name rf_data_phantom.mat in your directory for
the course.
Load the data into Matlab and look at the variables by writing whos.
The following variables are found in the file:
| Variable name |
Content |
Unit |
| RFdata |
The measured RF data from the transducer after TGC amplification.
The matrix contains one column for each of the lines in the image.
The data is 16 bits, two-complements integers, where a full range value
of 32768 corresponds to 2 volts. |
v |
| fs |
Sampling frequency |
Hz |
| start_of_data |
Starting depth for data |
m |
| start_angle |
Starting angle for data |
rad |
| angle |
Angle between the different RF lines |
rad |
| c_sound |
Speed of sound |
m/s |
| roc |
Radius of curvature. Distance from (0,0) to the patient side of the transducer. |
m |
The variables are defined in the figure below:
Plot the values for the center RF line in the image with the correct
depth scaling on the x-axis. Use the sampling frequency, start
depth and speed of sound for generating the correct depth values. Note
that the rf_data is int16 and should be converted to double before they can
be used.
-
Find the envelope of the data by making a Hilbert transform on each
of the RF signals (see help hilbert). Check the envelope by plotting
both a single RF signal and the envelope on top of each other.
-
Compress the envelope data to have a dynamic range of 60 dB and scale
it to lie from 0 to 127 for a colormap with 128 gray level values.
-
Download the display programs (fast_int_uint32.mexa64 for Linux,
fast_int_uint32.mexmaci64 in for_mac_matlab_2017 for Mac,
or fast_int_unit32.mexw64 for windows, and
make_interpolation.m, make_tables.m) from the
zip filke:
https://courses.healthtech.dtu.dk/22485/exercises/exercise1/programs/interpolation_programs.zip
and put them in the short course directory. Use the routines make_tables
and make_interpolation to perform the polar to rectangular
interpolation of the data. Write help make_tables and help make_interpolation
to find out how the routines are used.
-
Make the interpolation for all image files 8820e_B_mode_invivo_frame_no_<XX> where <XX> is
from 1 to 66. Each files houses an image like the one described
above. All files are found in the zip-archive:
in_vivo_data.zip.
The images can be combined into a movie by the Matlab commands avifile and addframe
(see the Matlab help for more information). The movie can be played by the
command movie. The frame rate i 22 fr/s.
|